Profile
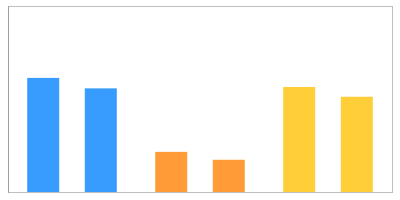
Quantities
The table shows normalized volumes (%Vol) of the visible expression profile for spot ID '13721' on gel image 'control_01'.
Ratios and p-values (using t-Tests) are computed relative to group 'control'.
| control | 1 min | 10 min | ||||
|---|---|---|---|---|---|---|
| control_01 | control_02 | 1min_01 | 1min_02 | 10min_01 | 10min_02 | |

|

|

|

|

|
|
|
| ID | 13721 | 13721 | 13721 | 13721 | 13721 | 13721 |
| Label | Eno | (no label) | (no label) | (no label) | (no label) | (no label) |
| X | 772 | 711 | 697 | 673 | 691 | 711 |
| Y | 343 | 323 | 342 | 315 | 343 | 347 |
| %V | 1.2268 | 1.1145 | 0.4316 | 0.3466 | 1.1291 | 1.0240 |
| Ratio | - | 0.332 | 0.920 | |||
| p-value | - | 0.008 | 0.346 | |||
| RSD | 4.795 | 10.914 | 4.882 | |||
| mean | 1.171 | 0.389 | 1.077 | |||
| median | 1.171 | 0.389 | 1.077 | |||
| maximum | 1.227 | 0.432 | 1.129 | |||
| minimum | 1.115 | 0.347 | 1.024 | |||
Labels
| Scout | Attribute | control | Fused Images |
|---|---|---|---|
| control_01 | Fused Image | ||
| Label name(s) | Eno | Eno | |
| Pi/MW Estimation | Pi | N.A. | N.A. |
| MW | N.A. | N.A. | |
| GenoList | Gene Size (bp) |
1290
|
1290
|
| Protein Size (aa) |
430
|
430
|
|
| Function |
|
|
|
| Uniprot | Accession |
P37869
|
|
| Amino Acids |
430
|
|
|
| Function |
Catalyzes the reversible conversion of 2-phosphoglycerate into phosphoenolpyruvate. It is essential for the degradation of carbohydrates via glycolysis.
|
|
|
| Gene name |
eno
|
|
|
| Gene Ontology |
|
|
|
| Isoelectric Point |
4.4037
|
|
|
| Keywords |
|
||
| Molecular Weight |
46581.39
|
|
|
| NCBI Taxonomy |
224308
|
|
|
| Organism |
Bacillus subtilis (strain 168)
|
|
|
| Original label name |
Eno
|
|
|
| Protein name |
Enolase
|
|
|
| Query text |
Eno
|
|
|
| References |
|
||
| Sequence |
MPYIVDVYAREVLDSRGNPTVEVEVYTETGAFGRALVPSGASTGEYEAVELRDGDKDRYL
GKGVLTAVNNVNEIIAPELLGFDVTEQNAIDQLLIELDGTENKGKLGANAILGVSMACAR
AAADFLQIPLYQYLGGFNSKTLPVPMMNIVNGGEHADNNVDIQEFMIMPVGAPNFREALR
MGAQIFHSLKSVLSAKGLNTAVGDEGGFAPNLGSNEEALQTIVEAIEKAGFKPGEEVKLA
MDAASSEFYNKEDGKYHLSGEGVVKTSAEMVDWYEELVSKYPIISIEDGLDENDWEGHKL
LTERLGKKVQLVGDDLFVTNTKKLSEGIKNGVGNSILIKVNQIGTLTETFDAIEMAKRAG
YTAVISHRSGETEDSTIADIAVATNAGQIKTGAPSRTDRVAKYNQLLRIEDQLAETAQYH
GINSFYNLNK
|
|
|
| Title |
ENO_BACSU
|
|
|
| Uniprot entry |
|
Position on Gel Image 'control_01'